ASM1 to ADM1 Translator
Introduction
A link is required to translate between biological and physically- or chemically-mediated processes to develop whole-plant modeling of wastewater treatment. This model mediates the interaction between the Activated Sludge Model 1 (ASM1) and the Anaerobic Digester Model 1 (ADM1).
The model relies on the following key assumptions:
supports only liquid phase
supports only ASM1 to ADM1 translations
Degrees of Freedom
The translator degrees of freedom are the inlet feed state variables:
temperature
pressure
volumetric flowrate
solute compositions
cations
anions
Ports
This model provides two ports:
inlet
outlet
Sets
Description |
Symbol |
Indices |
|---|---|---|
Time |
\(t\) |
[0] |
Inlet/outlet |
\(x\) |
[‘in’, ‘out’] |
Phases |
\(p\) |
[‘Liq’] |
Inlet Components |
\(j\) |
[‘H2O’, ‘S_I’, ‘S_S’, ‘X_I’, ‘X_S’, ‘X_BH’, ‘X_BA’, ‘X_P’, ‘S_O’, ‘S_NO’, ‘S_NH’, ‘S_ND’, ‘X_ND’, ‘S_ALK’] |
Ion |
\(j\) |
[‘S_cat’, ‘S_an’] * |
Outlet Components |
\(j\) |
[‘H2O’, ‘S_su’, ‘S_aa’, ‘S_fa’, ‘S_va’, ‘S_bu’, ‘S_pro’, ‘S_ac’, ‘S_h2’, ‘S_ch4’, ‘S_IC’, ‘S_IN’, ‘S_I’, ‘X_c’, ‘X_ch’, ‘X_pr’, ‘X_li’, ‘X_su’, ‘X_aa’, ‘X_fa’, ‘X_c4’, ‘X_pro’, ‘X_ac’, ‘X_h2’, ‘X_I’, ‘S_cat’, ‘S_an’, ‘S_co2’] |
- Notes
* Ion” is a subset of “Component” and uses the same symbol j.
ASM1 Components
Additional documentation on the ASM1 property model can be found here: Activated Sludge Model 1 Documentation
Description |
Symbol |
Variable |
|---|---|---|
Soluble inert organic matter, S_I |
\(S_I\) |
S_I |
Readily biodegradable substrate, S_S |
\(S_S\) |
S_S |
Particulate inert organic matter, X_I |
\(X_I\) |
X_I |
Slowly biodegradable substrate, X_S |
\(X_S\) |
X_S |
Active heterotrophic biomass, X_BH |
\(X_{BH}\) |
X_BH |
Active autotrophic biomass, X_BA |
\(X_{BA}\) |
X_BA |
Particulate products arising from biomass decay, X_P |
\(X_P\) |
X_P |
Oxygen, S_O |
\(S_O\) |
S_O |
Nitrate and nitrite nitrogen, S_NO |
\(S_{NO}\) |
S_NO |
\({NH_{4}}^{+}\) + \(NH_{3}\) Nitrogen, S_NH |
\(S_{NH}\) |
S_NH |
Soluble biodegradable organic nitrogen, S_ND |
\(S_{ND}\) |
S_ND |
Particulate biodegradable organic nitrogen, X_ND |
\(X_{ND}\) |
X_ND |
Alkalinity, S_ALK |
\(S_{ALK}\) |
S_ALK |
ADM1 Components
Additional documentation on the ADM1 property model can be found here: Anaerobic Digestion Model 1 Documentation
Description |
Symbol |
Variable |
|---|---|---|
Monosaccharides, S_su |
\(S_{su}\) |
S_su |
Amino acids, S_aa |
\(S_{aa}\) |
S_aa |
Long chain fatty acids, S_fa |
\(S_{fa}\) |
S_fa |
Total valerate, S_va |
\(S_{va}\) |
S_va |
Total butyrate, S_bu |
\(S_{bu}\) |
S_bu |
Total propionate, S_pro |
\(S_{pro}\) |
S_pro |
Total acetate, S_ac |
\(S_{ac}\) |
S_ac |
Hydrogen gas, S_h2 |
\(S_{h2}\) |
S_h2 |
Methane gas, S_ch4 |
\(S_{ch4}\) |
S_ch4 |
Inorganic carbon, S_IC |
\(S_{IC}\) |
S_IC |
Inorganic nitrogen, S_IN |
\(S_{IN}\) |
S_IN |
Soluble inerts, S_I |
\(S_I\) |
S_I |
Composites, X_c |
\(X_c\) |
X_c |
Carbohydrates, X_ch |
\(X_{ch}\) |
X_ch |
Proteins, X_pr |
\(X_{pr}\) |
X_pr |
Lipids, X_li |
\(X_{li}\) |
X_li |
Sugar degraders, X_su |
\(X_{su}\) |
X_su |
Amino acid degraders, X_aa |
\(X_{aa}\) |
X_aa |
Long chain fatty acid (LCFA) degraders, X_fa |
\(X_{fa}\) |
X_fa |
Valerate and butyrate degraders, X_c4 |
\(X_{c4}\) |
X_c4 |
Propionate degraders, X_pro |
\(X_{pro}\) |
X_pro |
Acetate degraders, X_ac |
\(X_{ac}\) |
X_ac |
Hydrogen degraders, X_h2 |
\(X_{h2}\) |
X_h2 |
Particulate inerts, X_I |
\(X_I\) |
X_I |
Total cation equivalents concentration, S_cat |
\(S_{cat}\) |
S_cat |
Total anion equivalents concentration, S_an |
\(S_{an}\) |
S_an |
Carbon dioxide, S_co2 |
\(S_{co2}\) |
S_co2 |
NOTE: \(S_{h2}\) and \(S_{ch4}\) have vapor phase and liquid phase, \(S_{co2}\) only has vapor phase, and the other components only have liquid phase. The amount of \(CO_2\) dissolved in the liquid phase is equivalent to \(S_{IC} - S_{HCO3^{-}}\) .
Parameters
Description |
Symbol |
Parameter Name |
Value |
Units |
|---|---|---|---|---|
Nitrogen fraction in particulate products |
\(i_{xe}\) |
i_xe |
0.06 |
\(\text{dimensionless}\) |
Nitrogen fraction in biomass |
\(i_{xb}\) |
i_xb |
0.08 |
\(\text{dimensionless}\) |
Anaerobic degradable fraction of \(X_I\) and:math:X_P |
\(f_{xI}\) |
f_xI |
0.05 |
\(\text{dimensionless}\) |
Equations and Relationships
Description |
Equation |
|---|---|
Pressure balance |
\(P_{out} = P_{in}\) |
Temperature balance |
\(T_{out} = T_{in}\) |
Volumetric flow equality |
\(F_{out} = F_{in}\) |
COD Equations
The total incoming COD is reduced in a step-wise manner until the COD demand has been satisfied. The reduction is based on a hierarchy of ASM1 state variables such that \(S_s\) is reduced by the COD demand first. If there is insufficient \(S_s\) present, then \(S_s\) is reduced to zero and the remaining demand is subtracted from \(X_s\). If necessary, \(X_{BH}\) and \(X_{BA}\) may also need to be reduced.
Description |
Equation |
|---|---|
COD demand |
\(COD_{demand} = S_{o} + 2.86S_{NO}\) |
Readily biodegradable substrate remaining (step 1) |
\(S_{S, inter} = S_{S} - COD_{demand}\) |
Slowly biodegradable substrate remaining (step 2) |
\(X_{S, inter} = X_{S} - COD_{demand, 2}\) |
Active heterotrophic biomass remaining (step 3) |
\(X_{BH, inter} = X_{BH} - COD_{demand, 3}\) |
Active autotrophic biomass remaining (step 4) |
\(X_{BA, inter} = X_{BA} - COD_{demand, 4}\) |
Soluble COD |
\(COD_{s} = S_{I} + S_{S, inter}\) |
Particulate COD |
\(COD_{p} = X_{I} + X_{S, inter} + X_{BH, inter} + X_{BA, inter} + X_{P}\) |
Total COD |
\(COD_{t} = COD_{s} + COD_{p}\) |
TKN Equation
The Total incoming Kjeldahl nitrogen is calculated with components updated in the anaerobic environment.
Description |
Equation |
|---|---|
Total Kjeldahl nitrogen |
\(TKN = S_{NH} + S_{ND} + X_{ND} + i_{xb}(X_{BH, inter} + X_{BA, inter}) + i_{xe}(X_{I} + X_{P})\) |
\(S_{nd}\) and \(S_s\) Mapping Equations
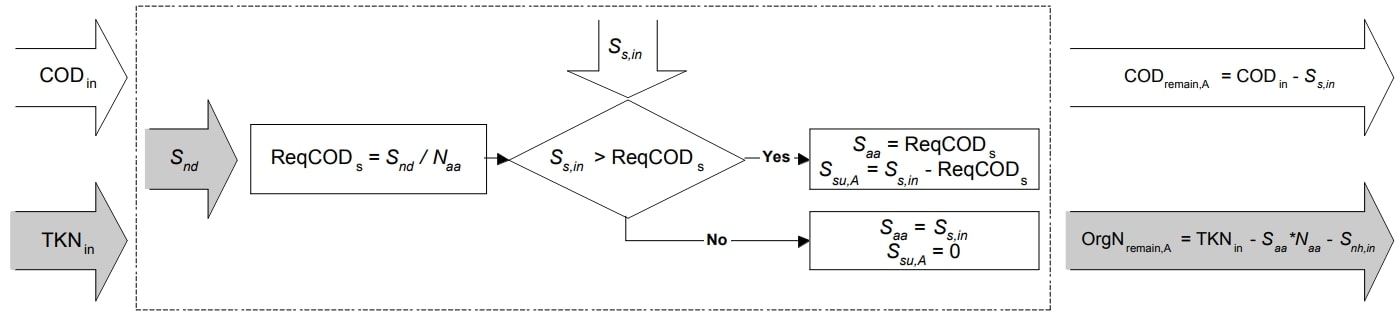
Figure 1. Schematic illustration of \(S_{nd}\) and \(S_s\) mapping (Copp et al. 2006)
Description |
Equation |
|---|---|
Required soluble COD |
\(ReqCOD_{s} = \frac{S_{ND}}{N_{aa} * 14}\) |
Amino acids mapping (if \(S_{S,inter} > ReqCOD_{s}\)) |
\(S_{aa} = ReqCOD_{s}\) |
Amino acids mapping (if \(S_{S,inter} ≤ ReqCOD_{s}\)) |
\(S_{aa} = S_{S, inter}\) |
Monosaccharides mapping step A (if \(S_{S,inter} > ReqCOD_{s}\)) |
\(S_{su, A} = S_{S, inter} - ReqCOD_{s}\) |
Monosaccharides mapping step A (if \(S_{S,inter} ≤ ReqCOD_{s}\)) |
\(S_{su, A} = 0\) |
COD remaining from step A |
\(COD_{remain, A} = COD_{t} - S_{S,inter}\) |
Organic nitrogen pool remaining from step A |
\(OrgN_{remain, A} = TKN - (S_{aa} * N_{aa} * 14) - S_{NH}\) |
Soluble Inert COD Mapping Equations
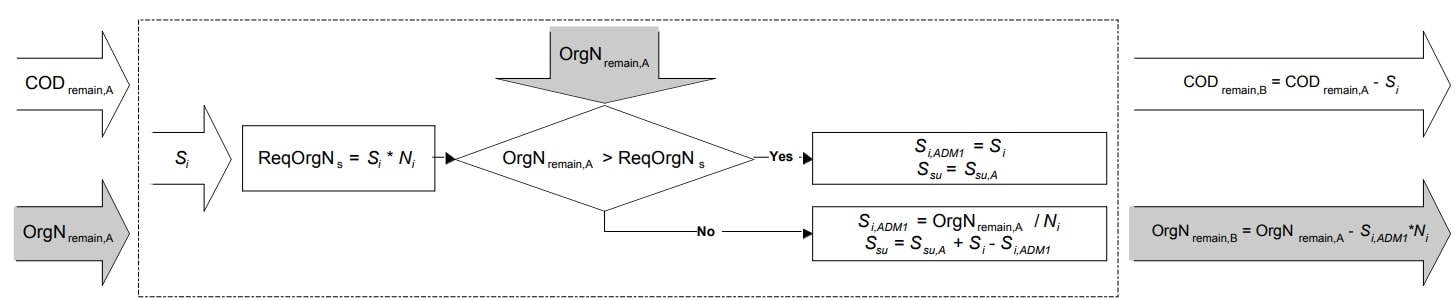
Figure 2. Schematic illustration of soluble inert COD mapping (Copp et al. 2006)
Description |
Equation |
|---|---|
Required soluble inert organic nitrogen |
\(OrgN_{s, req} = S_{I} * N_{I} * 14\) |
Soluble inert mapping step B (if \(OrgN_{remain, A} > OrgN_{s, req}\)) |
\(S_{I, ADM1} = S_{I}\) |
Soluble inert mapping step B (if \(OrgN_{remain, A} ≤ OrgN_{s, req}\)) |
\(S_{I, ADM1} = \frac{OrgN_{remain, A}}{N_{I} * 14}\) |
Monosaccharides mapping step B (if \(OrgN_{remain, A} > OrgN_{s, req}\)) |
\(S_{su} = S_{su, A}\) |
Monosaccharides mapping step B (if \(OrgN_{remain, A} ≤ OrgN_{s, req}\)) |
\(S_{su} = S_{su, A} + S_{I} - S_{I, ADM1}\) |
COD remaining from step B |
\(COD_{remain, B} = COD_{remain, A} - S_{I}\) |
Organic nitrogen pool remaining from step B |
\(OrgN_{remain, B} = OrgN_{remain, A} - (S_{I, ADM1} * N_{I} * 14)\) |
Particulate Inert COD Mapping Equations
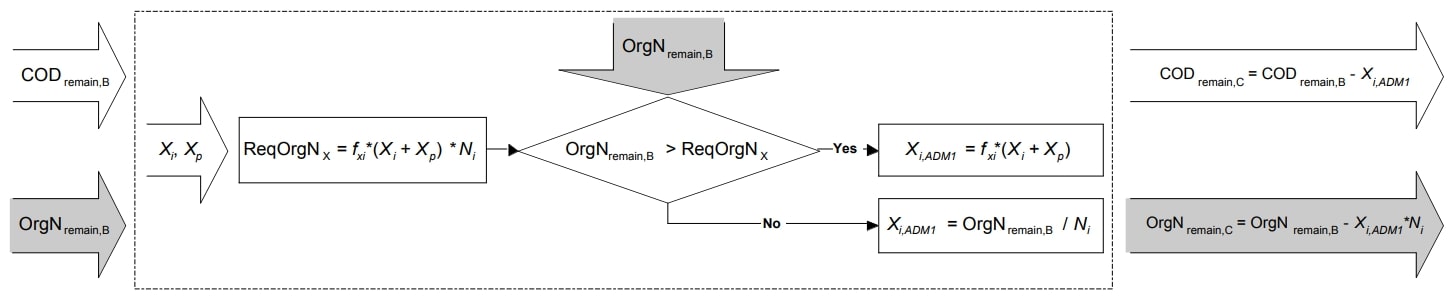
Figure 3. Schematic illustration of particulate inert COD mapping (Copp et al. 2006)
Description |
Equation |
|---|---|
Required particulate inert material |
\(OrgN_{x, req} = f_{xi} * (X_{P} + X_{I}) * N_{I} * 14\) |
Particulate inert mapping step C (if \(OrgN_{remain, B} > OrgN_{x, req}\)) |
\(X_{I, ADM1} = f_{xi} * (X_{P} + X_{I})\) |
Particulate inert mapping step C (if \(OrgN_{remain, B} ≤ OrgN_{x, req}\)) |
\(X_{I, ADM1} = \frac{OrgN_{remain, B}}{N_{I} * 14}\) |
COD remaining from step C |
\(COD_{remain, C} = COD_{remain, B} - X_{I, ADM1}\) |
Organic nitrogen pool remaining from step C |
\(OrgN_{remain, C} = OrgN_{remain, B} - (X_{I_ADM1} * N_{I} * 14)\) |
Final COD and TKN Mapping Equations
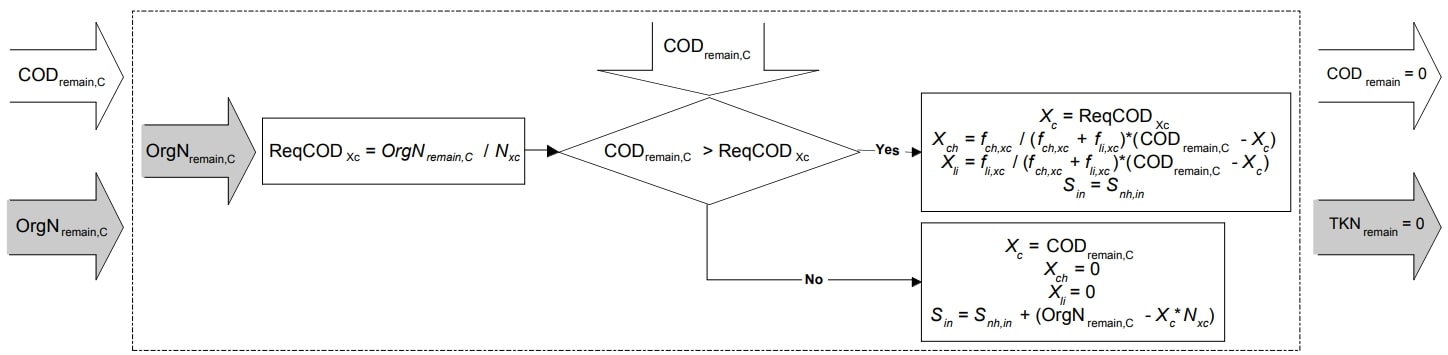
Figure 4. Schematic illustration of final COD and TKN mapping (Copp et al. 2006)
Description |
Equation |
|---|---|
Required soluble COD |
\(COD_{Xc, req} = \frac{OrgN_{remain, C}}{N_{xc} * 14}\) |
Composites mapping (if \(COD_{remain, C} > COD_{Xc, req}\)) |
\(X_{C} = COD_{Xc, req}\) |
Composites mapping (if \(COD_{remain, C} ≤ COD_{Xc, req}\)) |
\(X_{C} = COD_{remain, C}\) |
Carbohydrates mapping (if \(COD_{remain, C} > COD_{Xc, req}\)) |
\(X_{ch} = \frac{f_{ch, xc} (COD_{remain, C} - X_{C})}{f_{ch, xc} - f_{li, xc}}\) |
Carbohydrates mapping (if \(COD_{remain, C} ≤ COD_{Xc, req}\)) |
\(X_{ch} = 0\) |
Lipids mapping (if \(COD_{remain, C} > COD_{Xc, req}\)) |
\(X_{li} = \frac{f_{li, xc} (COD_{remain, C} - X_{C})}{f_{ch, xc} - f_{li, xc}}\) |
Lipdis mapping (if \(COD_{remain, C} ≤ COD_{Xc, req}\)) |
\(X_{li} = 0\) |
Inorganic nitrogen mapping (if \(COD_{remain, C} > COD_{Xc, req}\)) |
\(S_{IN} = S_{NH, in}\) |
Inorganic nitrogen mapping (if \(COD_{remain, C} ≤ COD_{Xc, req}\)) |
\(S_{IN} = S_{NH, in} + (OrgN_{remain, C} - X_{C} * N_{xc} * 14)\) |
Anions balance |
\(S_{an} = \frac{S_{IN}}{14}\) |
Cations balance |
\(S_{cat} = \frac{S_{IC}}{12}\) |
Classes
- class watertap.unit_models.translators.translator_asm1_adm1.TranslatorDataASM1ADM1(component)[source]
Translator block representing the ASM1/ADM1 interface
- build()[source]
Begin building model. :param None:
- Returns:
None
- initialize_build(state_args_in=None, state_args_out=None, outlvl=0, solver=None, optarg=None)[source]
This method calls the initialization method of the state blocks.
- Keyword Arguments:
state_args_in – a dict of arguments to be passed to the inlet property package (to provide an initial state for initialization (see documentation of the specific property package) (default = None).
state_args_out – a dict of arguments to be passed to the outlet property package (to provide an initial state for initialization (see documentation of the specific property package) (default = None).
outlvl – sets output level of initialization routine
optarg – solver options dictionary object (default=None, use default solver options)
solver – str indicating which solver to use during initialization (default = None, use default solver)
- Returns:
None
References
[1] Copp J. and Jeppsson, U., Rosen, C., 2006. Towards an ASM1 - ADM1 State Variable Interface for Plant-Wide Wastewater Treatment Modeling. Proceedings of the Water Environment Federation, 2003, pp 498-510. https://www.accesswater.org/publications/proceedings/-290550/towards-an-asm1—adm1-state-variable-interface-for-plant-wide-wastewater-treatment-modeling